Return to the Course Home Page
There is no project to clone for this week; you can do all work in your own project
Sequencing and Mapping
A/Prof Olin Silander
Purpose
Introduction
Background
SARS-CoV-2 Genome Sequencing
Illumina
PacBio
Oxford Nanopore
Software Management
Conda Installation
Naming Conventions
Software Installation
SARS-CoV-2 Genome Sequencing
Our Data
Critically Evaluating Your Data
Plotting the Data more Deliberately in R
Portfolio Analysis
Purpose
- To learn how to use a package manager to install software for use on the command line.
- To understand the advantages and disadvantages of using two of the three most common types of NGS sequencing data.
- To be able to visualise the differences in these NGS data types.
- To discuss what the possible applications are for this type of NGS data.
Introduction
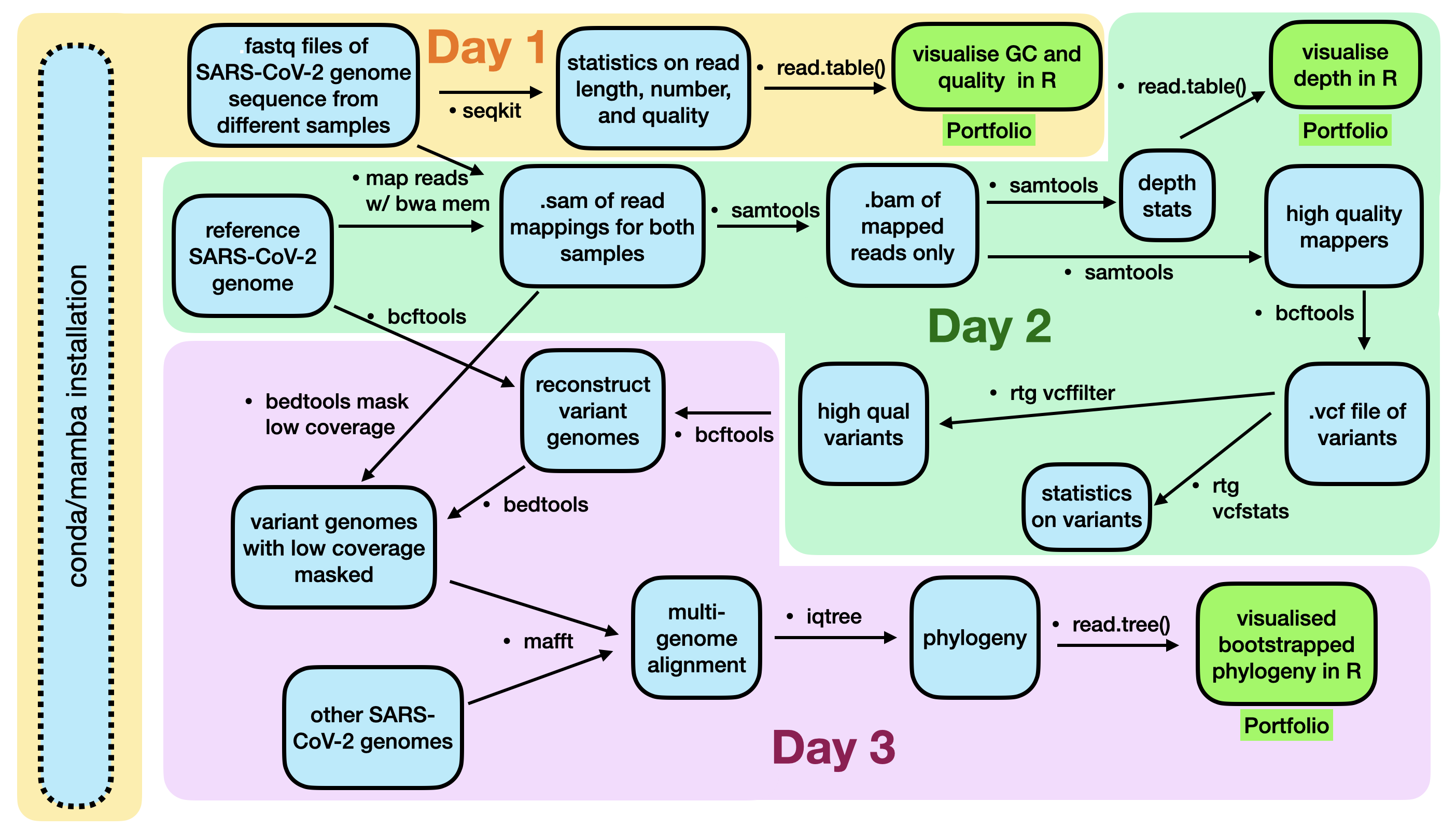
The data we will investigate today is from publicly available SARS-Cov-2 genome sequences. Over the next two weeks. you will learn how to use this data to find specific mutations that new SARS-CoV-2 strains have, how to place these strains in an evolutionary context, and how to visualise this context.
A flowchart of the process we will follow is below. It is unlikely that you will understand all of these steps right now, but hopefully after the lecture content and labs in the next three weeks, this will become more clear.
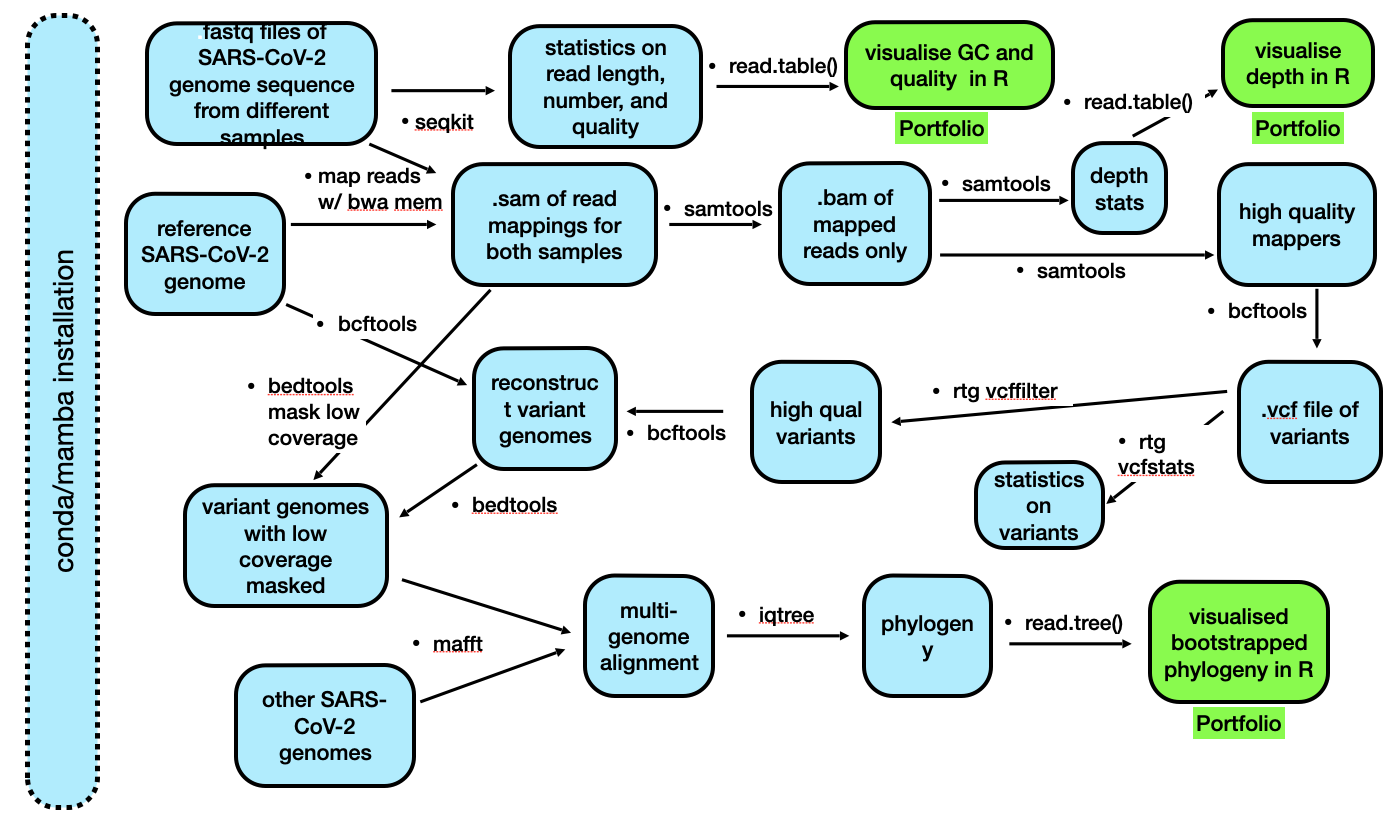
Background
Soon after the birth of Next Generation Sequencing in 2005 (or so), the technology rapidly proliferated into a number of different platforms (e.g. 454, IonTorrent, Helicos, and others). Over the last decade and a half, companies came and went, and currently there are three dominant NGS sequencing platforms: Illumina, which dominates the market; PacBio; and Oxford Nanopore. These three technologies differ considerably in their methodologies. For all three, sequencing output ranges from tens of gigabases (billions of nucleotides) to terabases (trillions of nucleotides), depending on the specific platform (e.g. Illumina MiSeq, Illumina NovaSeq, Oxford Nanopore MinION, Oxford Nanopore P2, etc.). However, these are not the only methods available, and recently a number of other possibly disruptive technologies have come onto the scene (see below). Lest we forget, when Fred Sanger first started sequencing, he was averaging one base pair per day at his peak.
Illumina

Illumina sequencing relies on sequencing-by-synthesis, a process in which hundreds of millions of single DNA molecules deposited onto a patterned flowcell. These single molecules are then multiplied into hundreds of millions of “clusters”, each consisting of thousands of clonal molecules (each derived from a single molecule). Each cluster is sequenced by (1)incorporating a fluorescent nucleotide (the same nucleotide will be incorporated into all molecules in a cluster), (2) exciting the fluorescence of the incorporated nucleotides, (3) and taking a picture of the cluster’s fluorescence. The fluorescent moiety (big word) is then cleaved and the next nucleotide is incorporated. Review the method here. Read lengths for Illumina range between 75 bp and 300 bp, and are of very high quality (i.e. the sequence of base pairs is almost certainly correct, with an error rate of approximately 1 in 10,000).
PacBio
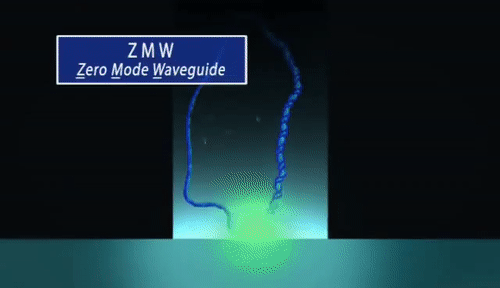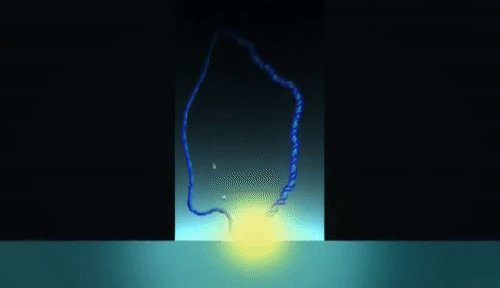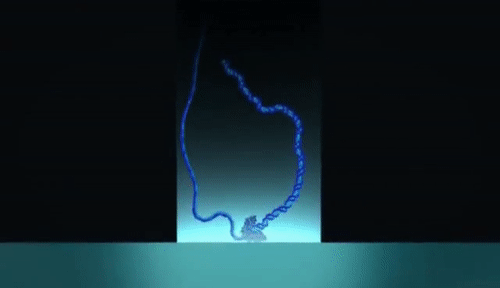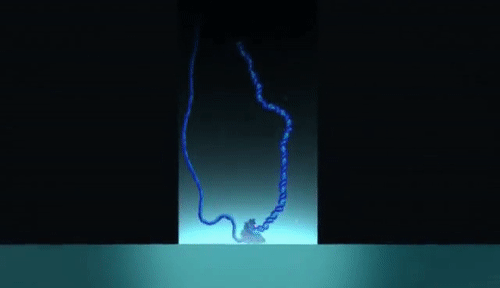
PacBio sequencing relies on imaging fluorescent nucleotides as they are incorporated into single DNA molecules using zero-mode-waveguides (“the worlds smallest microscope”). This is fundamentally different from Illumina in that the pictures that are taken during sequencing are of single DNA molecules Review the method here. Read lengths for PacBio range from 1000 bp to 30 kilobase pairs, and range in quality from very low (15% error rate) to very high (1 in 100,000 or lower error rate).
Oxford Nanopore
Oxford Nanopore sequencing relies on sensing current changes in a pore as a single DNA or RNA molecule is passed through a pore (a protein taken from E. coli). This differs fundamentally from both the above methods as it does not rely on taking pictures. Review the method here. Read lengths for Oxford Nanopore are essentially unlimited (e.g. 1 megabase pair), and are of medium quality, with an error rate of approximately 1 in 100.
MGI / BGI
MGI is a relatively new technology that relies on creating DNA “nanoballs” (DNB). After these are made, they are deposited on a patterned flow cell (as Illumina is), fluorescent nucleotides are added, and a picture of the flow cell is taken to determine the colour of the nucleotide that has been incorporated. For a video of this process, see here. This technology is very very similar to Illumina’s, with similar read lengths and accuracy.
ElementBio Aviti and Ultima Genomics
The Element Aviti is one of the newer technologies that offer high throughput sequencing. It is so new that I have not figured out exactly how it works, despite multiple different attempts. There is some rolling circle stuff (like MGI), some polony stuff, some labelled (fluorescent) nucleotides, and some non-fluorescent nucleotides. It is similar to - if not quite identical - “sequencing-by-synthesis.” Again, this has similar read lengths and accuracy as Illumina (and in fact offer a kit to convert Illumina libraries so they are compatible with Aviti).
Ultima is the most recent big entry into the NGS market, and promises by a considerable margin, the cheapest human genome ($100). Ultima also uses “sequencing by synthesis” (mostly), as well as a CD-like surface that allows smaller amounts of reagents per sequencing run.
Which platforms matter now and in the future?
It’s difficult to know what the sequencing landscape will look like in five years. However, current interests from people involved in sequencing may give us some idea. Albert Vilella recently posted a Twitter poll: Which new sequencing platform/company do you find most interesting?. See the bottom of this page for a complete summary of all current NGS sequencing platforms and their costs.
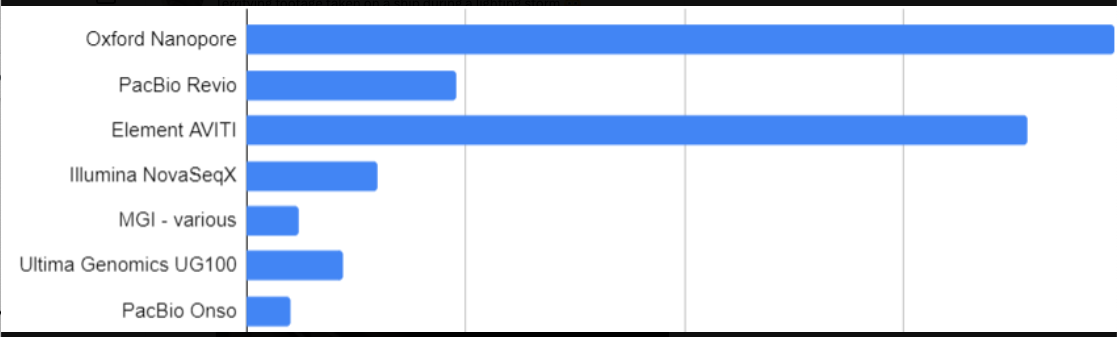
Looks like there’s a lot of interest in Oxford Nanopore and Element
Today
Note: we will refer to any DNA sequence data from an NGS platform as a “read”.
Today we will deal with DNA sequence data from two of the most widely-available technologies, Illumina and Oxford Nanopore. The primary difference between these two technolgies is that Illumina provides short, highly accurate reads using a very expensive machine (~ $1 million USD), while Oxford Nanopore provides long, less accurate reads using a very cheap machine (~ $1000 USD). As you can imagine, these characteristics provide different advantages.
Oxford Nanopore and Illumina also differ in other ways, but we will not discuss those in detail today. Perhaps the primary difference is that Oxford Nanopore sequences the original molecules of DNA and RNA with all their various modifications (cytosine methylation, adenine methylation, and others), whereas Illumia sequences copies of DNA only.

Oxford Nanopore - the original DNA in all its glory
First things first
We will soon get to the actual DNA sequence data. But to deal with the data, you will need a slew of new software. Thus, you must be introduced to methods for managing that software. Let’s begin there.
Software Management
We need software to be able to process all of this data. Until now (other than Linux, the operating system itself), you have used the pre-installed statistical programming software, R. However, we need additional software to process DNA sequence data.
Note: Here, rather than just “software”, we will refer to these computer programs as “software packages”, as one piece of “software” may contain several pieces of related software, hence software package (you have seen the phrase software packages before when installing new programs in
R).
As you are probably aware, software packages are sets of tools that have been developed to perform specific jobs, or are used to implement specific methods. Your general view of a software package may be something like Excel or Chrome or TikTok. More fundamentally, a software package is simply a group of computer programs used to perform a specific task. In bioinformatics, for example, this could be a set of programs (individual computer programs written in a language such as python) telling the computer how to interpret and display the quality scores from a .fastq file.
However, software packages and tools often have dependencies, which are other pieces of software that are necessary to run the software you would like to install. For example, to use Instagram, you also need software that controls your phone’s camera. This reliance of Instagram on camera-controlling software is known as a dependency. Importantly, software like Instagram is designed to be user-friendly, and during installation will usually check whether such camera-controlling software exists, and if it does not, may try to install it.
Software dependencies are real and precarious, especially in bioinformatics (credit: xkcd)
Despite the existence of dependencies, many bioinformatics software programs (most of which are written by academic-oriented computational biologists – or worse, plain-old biologists) do not check for dependencies. This can create significant issues if you try to run a piece of software but are missing other software that it needs to run. To make sure that we resolve all these dependency issues when we install new software, we will use a package management system. This management system is called conda, and it is perhaps the most common package manager used in bioinformatics. It considerably simplifies the process of installing software, meaning that you will not need to find software websites, download multiple files, unzip files, find compatible files for your operating system, etc.

You will never escape them
Conda Installation
As with any software, the first thing we need to do is install the package manager itself. The installation of this tool is perhaps the most complicated installation we will do in this course, as we cannot use conda to install itself (and I have not pre-installed it on your system). However, after the installation of conda, your life will become far easier (well, in terms of analysing biological data) and you will be on your way to becoming a seasoned bioinformatician.
First, I need to post a reminder – as we will be operating mostly on the command line, you must never forget tab-complete.

Tab complete will solve all your problems
You must also never forget the up arrow.

Every seasoned bioinformatician uses it.
Second, try to follow the instructions exactly today, and whatever you do don’t click here as it will delete all your files. If you get an error or warning of any sort, go back and make sure you have followed the instructions. If you continue to get the error, then it could be my fault.
Throughout today, commands will appear in grey boxes, sometimes with plentiful comments above. Please read the comments to understnad what to do, and what the command is doing.
Good. Now, we download conda.
Navigate to the command line tab on your RStudio window (“Terminal”). This is on the top of the R window. Make sure you are in your /cloud/project/ directory.
# Download the latest conda installer.
# We cry because we can't use tab-complete here as
# the file does not yet exist on our computers.
# you should be able to copy the line below and
# paste it on the command line
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
Explanation: wget is a program that is used to transfer data to or from a server. Thus, this command is simply using wget program to find the file at the location indicated and then download it.
The file you have downloaded (with the extension .sh) is a bash file, which is usually run using the command line program bash. As you know, noting the extension of a file can be very helpful in figuring out what is in it, or what it does. For example, you should never end a bash file with .txt as that suggests it is a simple text file, when in fact it is not. Similarly, you would never end a Microsoft Word file with .xlsx, you would end it with .doc or .docx. And if you do find a file with the suffix .sh you can guess it’s a bash file and use bash to run it.
Naming Conventions
One important aspect of organising files and directories (folders) is naming convention. When working on the command line, your life will become considerably easier if you avoid using spaces in your files and directory names. Thus, never name your file my awesome file.txt. Instead, name it my_awesome_file.txt (“snake case”), or myAwesomeFile.txt (“camel case”) or my-awesome-file.txt (“kebab case”) or my.awesome.file.txt and probably not MY_AWESOME_FILE.txt (“screaming snake case”) or MY-AWESOME-FILE.txt (“spicy kebab case” (okay I might’ve made that up)).
You should pick one of these at the start of the course, and stick to that format throughout the course (i.e. camel case, or kebab case, etc.) Know that there is no clear-cut best case convention. I usually use snake case, but not always - kebab case requires one less keystroke than snake case so I sometimes use that. And using a . means that your file names will only ever have one type of non-word character, so it’s less to remember. But, do as I say not as I do and always use the same convention. Last, you should almost never begin a file or directory name with a . (e.g. .my-awesome-file.txt) as this will make it a hidden file.

Please be consistent with your file names
As I pointed out above and will re-emphasise here, the second thing to pay attention to when naming files is the extension or suffix. For example text files are usually named with the extension .txt. Often, but not always, file extensions have three characters. Some well-known exceptions are .html, .docx, .xlsx, and the perhaps not standard .jpeg. In this course, we will run into a wide variety of files with a wide variety of extensions, for example .fastq, .sam, .bam, .txt, .sh, .fasta, .html, .gbk, .bai, .py, .r (sometimes .R), .gz, .aln, .tre, .phy, .vcf, .bcf, and many more! Hopefully at the conclusion of this Semester you will be familiar with all of these.
Aside: if you are ever writing the date, use the format YYYYMMDD. That way, when you sort files by name, they will also be sorted by date (e.g. if you know that you made one set of data before another, it will be easier to find).
Finally, there are certain characters that you should always avoid when naming files and folders. Besides spaces, these are (not necessarily exhaustive):
: ; ` " ' \ / ! @ # $ % ^ & * ( ) + , ? [ ] { } | > <
Look closely above and you will note that several characters above are a different colour (e.g. “;”) - that’s because the bash interpreter used to render this webpage thinks it’s a special character.
Back to the Topic at Hand - Conda (and Mamba), a Package Manager
Let’s now actually install conda (in our case we install a miniature version of it with less bloat, miniconda).
Warning: Be careful when using rm in the following command. (Why? What does rm do?)
# Run the conda installer
# Note: now you can use tab-complete.
# During installation. You will need to
# press enter and the spacebar several
# times at the --More-- prompt, and
# type "yes" each time it prompts you to. It should
# be readily apparent where to do this:
# "Miniconda3 will now be installed into this location:" yes
# "Do you wish the installer to initialize Miniconda3
# by running conda init?" yes
bash Miniconda3-latest-Linux-x86_64.sh
# delete the installer after successful run
rm Miniconda3-latest-Linux-x86_64.sh
IMPORTANT
Conda may not behave quite as it should in this cloud-based platform. Try typing conda --help. If there is an error, then to use conda we will need to addjust our $PATH variable.1 You do this by typing the following at the terminal (simply copy-paste) the entire line below:
# Make sure you type this EXACTLY, including the $ !!!
export PATH="$HOME/miniconda3/bin:$PATH"
If you keep your terminal session active (i.e. don’t close it) then you should be able to use conda for the rest of the Semester in the terminal. If you do close the terminal, repeat the above step.
Now, you should be able to use the conda command. Again, one useful way to check that conda (or most other command line programs) is working is to ask the program for help. This is almost always done by typing --help or -h after the command. For example try:
conda --help
This should bring up a list of sub-commands that conda can do (assuming you have installed it correctly). If this does not work, ask someone for help (lecturer, demonstrator, or classmate). Note that this is different from the R help command.
Also note that for most command line programs, options are specified using one dash (e.g. -h) or two dashes (e.g. --help). It’s sometimes hard to know which one though.

Oh man, which is it.
Faster Management
Over the years, the conda ecosystem has gotten so large that it is slow and sometimes painful to navigate. For this reason, we will use a faster manager, mamba. mamba will allow faster searching of conda channels as it works in parallel and uses C++. Install mamba using the following syntax:
# don't worry about exactly what is happening here
conda install mamba -n base -c conda-forge
This will likely “redline” your RAM (visible in the top right corner of your screen as a little odometer). However, once installation occurs, the RAM should decrease. If the install fails, let someone know.
As you can see, we have used conda only to be able to install mamba. From now on, use mamba to install all programs.
Software Installation
mamba installs software packages using what is called a recipe, and these recipes are contained in places called channels. Most recipes for bioinformatic software are contained in the bioconda channel, which currently has recipes for more than 7000 software packages.
Let’s try to install some software packages now.
One piece of software we will need allows us to figure out where a certain sequence of DNA is in a genome (this type of software can be referred to as a “mapper” because it “maps” one sequence of DNA to another sequence of DNA). We will explore why you might want to do this later. The software we will use is minimap2. This installation process is relatively simple:
# Below we specify which channel we would like mamba
# to look in to find the minimap2 recipe.
# This is done using the -c option
mamba install -c bioconda minimap2
mamba will sit around looking for the recipe for a minute, and then it should ask you whether you want to install the software. Simply press enter or type Y and press enter.
Congrats! Your first command line installation of a real software package!
Let’s now get to the task at hand for today: analyzing DNA sequences from SARS-CoV-2.
SARS-CoV-2 Genome Sequencing
In the past 3.5 years, SARS-CoV-2 (the causative agent of COVID-19) has become one of the most extensivley sequenced organisms on earth, with well over ten million whole genome sequences available, and as of a year ago, two million sequence by the UK alone. SARS-CoV-2 genome sequencing is performed for two primary reasons: (1) to track the emergence of new and possibly more virulent variants, and (2) to track transmission between people. It is this second application that was used extensively in New Zealand early in the pandemic, in constrast to most other countries.
QUESTION
- Why was it possible to use SARS-CoV-2 genome sequencing for transmission tracking in New Zealand but not in most other countries?
Please look over this paper here, especially figures 3 and 4; and this paper here (how many of those outbreaks do you remember?) for some applications of SARS-CoV-2 genome sequencing data in New Zealand. Both of these papers are required reading and may appear in your tests.
Our Data
There are several methods used to sequence SARS-CoV-2, but perhaps the most common is via amplicon panels, in which PCR is used to amplify the entire genome in short pieces, which are then sequenced. The four most common methods are listed here. The “xGen SARS-CoV-2 Midnight Amplicon Panel” 😜 is the method used to produce the data we will explore here.
The NGS methods that produced today’s data (sequence data from different SARS-CoV-2 isolates) are Illumina and Oxford Nanopore. The format of the sequence data is fastq. Remember that the fastq format specifies a name for each sequence, the sequence itself (i.e. order of basepairs), and the quality of each basepair (i.e. how certain the sequencing machine is that it is giving you the correct base). Review fastq format here.
The sequence data are available as a normal tarball here as a tar.gz file. Do not download the data yet.
To download the data, first make sure you are in your /cloud/project directory. Second, make a new directory, perhaps covid/, and change into that directory. Third, copy the link address (right click on the link and scroll to Copy Link Address). Finally, download the files using wget:
# try downloading the tar file first.
# The link you copied shouldbe something like:
# "./data"
wget tarball-link-address-you-just-copied
You should see a rapid animated arrow tracking the download.
This should result in a ~17Mb tar ball in your directory. If it does not, then try using this link. If you use this link you will have to do the following:
# The downloaded file will have an odd extension.
# you will need to change it using mv.
# TAB-COMPLETE the first filename below to do this
mv sequence-files.tar?raw=true sequence-files.tar
Once you have downloaded the data, you will need to unpack it. Use tar to do this:
# expand the tar ball
tar -xvf sequence-files.tar
This should result in a single directory, sequence-files, containing four compressed fastq files (fastq.gz). Two of these are Illumina files, and two are Oxford Nanopore.
Nopte that the Illumina data are paired end, so there are two files)2, but these are from the same SARS-CoV-2 viral isolate.
Quick note: here and throughout the lab sessions I will often refer to certain files or directories as myfile.txt or mydir/. This does not mean that you should use this name, or that this file or directory exists for you. Rather, you should replace this name with a filename that does exist or which you do want to analyse or which is nicely descriptive. For example, intead of a covid/data/ directory above, you could make a directory called scv/sequence_data/. Feel free to change the name now if you like (hint: use mv).
After downloading all four files, you will have all the DNA sequence data that we will use today. If you have done this correctly, you should be able to list the files from the command line. Please do that now. There are several ways to do this. In this case you need to know (1) whether there are files in the directory, and (2) whether there is anything in the files (and preferably how much data is in the files). If you are not familiar with how you might do this, click here to see the man pages on ls.
QUESTIONS
- Are all four files present?
- How big are they?
- Are you sure they all sitting in the
/datadirectory that is sitting within your/cloud/project/directory? - What does the
.gzat the end of the file names indicate about the type of file it is? - How can you view the contents of files that have a
.gzsuffix (for example, what do you need to do first before viewing)?
Organization (optional but useful and easy)
Things will start to get a little crazy as we get more data - new directories, new files, too many ls commands. Let’s see if we can look inside this maze of files in a more accessible way. How? tree. Let’s install tree:
# here we actually change our channel
# i.e. it's no longer bioconda
# But we still use the -c option to
# specify another channel
mamba install -c conda-forge tree
Now we can use our tree command to see what is where and how it’s organised:
# We use the -L option, the -h option, and the --du option
# First head back up a directory (it does not matter where
# you are, just that you have at least one directory
# or file below you)
cd ..
# Then run tree
tree -L 2 --du -h
Nice.
Check out the tree command using the --help subcommand. What do the -L --du -h options do?
Critically Evaluating Your Data
Making Good Use of Summary Statistics
Next, let’s next look quickly inside the sequence data files. However, we don’t want to open them up - they’re quite large (well, not that large for genome sequence data). We will use the simple terminal command you know well, head (you could also use less or tail. You have encountered all of them previously):
# Here we first have to use zcat, not head directly, or cat,
# as the files are zipped.
# We then pipe | the result to head (remember, head is the command used to
# look at the first few lines of a file - by default, ten lines).
# You've also encountered the pipe before - it takes the output of one command
# and feeds it to another.
zcat choose_one_fastq_file_to_look_at.fastq.gz | head
For completeness and carefulness, we do it for all four files. Delete any suspicious sequence files from your directory and note to yourself the importance of looking critically at your data.
Once we have the data and have decided that it looks as we expect (or does it? 🤔), the first thing we will do is get some summary statistics (all good data science and bioinformatics and, indeed, any science should begin with actually looking at the data). Luckily, there are a number of other pieces of software that have been written to do this, so we will not need to re-invent the wheel. Today we will use two pieces of software. The first is seqkit, a blazingly fast and flexible piece of software. Install:
# Below we use mamba (of course) and
# tell mamba which *channel* to look in
# for the recipe using the -c option
mamba install -c bioconda seqkit
If your command does not work, let a lecturer, demonstrator, or classmate know.
Summary Stats with seqkit
Now let’s use seqkit (“sequence kit”) first. Type seqkit --help to make sure it’s working. No errors? If you have an error, ask for help. First, some simple statistics about your read files:
# Some simple statistics about your files
# these will list the numbers in basepairs
# e.g. total bp, etc.
# here, stats is a *subcommand* of seqkit
# this also means it won't tab-complete (as it is a subcommand)
seqkit stats *fastq.gz
QUESTIONS
- What does
*fastq.gzmean in the above command (i.e. what is the*doing)? - How many reads are in each file?
- How many total base pairs of data are in each file?
- How do the average read lengths differ between your sequencing files?
# Maybe a few more seqkit stats using the -a option. Remember the
# *correct name* of the program and you know that
# gzipped files don't end in ".gx" so
# don't copy-paste
seqklt stats -a *fastq.gx
Let’s look at whole distributions of read lengths instead of just the average read length for all read:
# A histogram of read lengths.
# We use the *watch* subcommand and the --bins option to clean up the
# histograms a tiny bit. The --bins options tells the program
# how many different histogram bins you want (here, I chose 15)
# If you want, you can leave the --bins 15 part of the command out.
# Note that the fastq.gz file name below is not the same as yours!
# The subcommand is "watch" not "witch", so copy-paste won't be easy
seqkit witch --bins 15 choose_one_fastq_file_to_plot.fastq.gz
Use this seqkit watch command for all of your sequencing files. You can also try the --log option if you want (what does this option do?). Remember, the up-arrow and tab-complete are your friends. --help is your other friend, e.g. seqkit watch --help.
QUESTION
- How do the sequencing files differ in the distributions of read lengths?
It is also possible to make a simple plot of the average quality of each read. In this case, quality of a base (A,C,G,T) in a read refers to the likelihood that the base is correct. See the explanation of quality scores here. Recall that Illumina and Oxford Nanopore data differ in their read accuracy, and thus quality. Which technology has higher accuracy? Go ahead and plot the quality scores. Here we also use seqkit.
# Below we add a new argument, --fields, to specify which
# read characteristic we would like to plot. Above seqkit used
# the default field for watch, MeanLength. Now we can get MeanQual.
# We leave the --bins option in (you don't have to).
# When you leave in --bins you need to specify a number (I chose 15)
seqklt wutch --fielbs MoanQual --bins 15 choose_one_fastq_file_to_plot.fastq.gz
Any errors? The program is “seqkit”, the subcommand is watch, the argument is fields, and the read characteristic is MeanQual.
Do this for both the Oxford Nanopore and Illumina reads.
QUESTIONS
- If a base has a quality score of 10, what is the likelihood that it is the correct base? See if you can find this out using Google, or calculate it if you remember how to convert q-score to accuracy.
- What if a base has a quality score of 25? Again, see if you can search for this answer or calculate it.
- How do the Illumina and Oxford Nanopore sequencing files differ in the distributions of quality scores?
- What are the highest average read qualities for the Oxford Nanopore reads (approximately)?
- For an Oxford Nanopore read with a “high” average quality, what fraction of bases do you expect to be correct?
Summary Stats with fastp
There are a number of other ways to look at your sequence data, and some of them are far more aesthetically pleasing than seqkit. One of these is fastp, which provides interactive reports via html. fastp is somewhat fiddly to get working on your versions of RStudioCloud, so instead I provide an example report here. Download this by clicking on the link, and then expand (untar) the files by double clicking the downloaded tar file. This will open a folder called “fastp”. Double click on the html file, and this should open in your default browser. Please look at this report now.
QUESTIONS
- What percentage of reads were scored as having low quality?
- Are any specific bases generally of lower quality (e.g. A, C, G, T)?
- What text is at the very bottom of the web page? Why is this useful?
Getting Tabulated Data to Plot in R

Me explaining the joy of learning R credit: @evornithology Twitter
In the fastp report we have several useful statistics, such as the mean read quality for each base over the length of the reads, and the fraction of bases at each position that are A, C, G, or T. However, there are additional important statistics that are not provided. Some of these we saw with the seqkit tool kit. But these plots were not aesthetically pleasing, and in some cases difficult to interpret. For that reason, we are going to replot some data, as well as additional data, using our familiar plotting software, R.
To begin, we make some files with the new data we would like. In this case, we will use seqkit again. First, some data on the distribution of lengths and quality per sequence (rather than per position). Do this for your untrimmed and trimmed Oxford Nanopore data (Montana):
#let's try seqkit fx2tab ("fastx format to tab format")
# fx2tab is an odd-looking command, but it is the correct command.
# Again, fx2tab is a *subcommand* of seqkit and will not tab-complete
seqkit fx2tab --help
# -q and -l options give the length and quality for each sequence.
# -n supresses the output of the sequences and qualities for *every bp*
# of each read
# note that here we can squish them into a single option, -qln rather
# than -q -l -n (also possible)
# so here we will have a file with three columns:
# the NAME, the LENGTH, and the QUALITY
seqkit fx2tab -qln myseqs.fastq.gz
Oops. Did you forget that seqkit outputs to standard out? You have to use the redirect > to put it into a file.
# Please do not redirect this into mydata.txt
# Rather, name the file something sensible.
seqkit fx2tab -qln myseqs.fastq.gz > mydata.txt
QUESTION
Why have I used .txt at the end of the example file above?
Plotting the Data more Deliberately in R
Now we have to think.
- What types of plots are we interested in?
- What sort of information would we like to see?
- What sort of information is it possible for us to see?
- What sort of story would we like our plots to tell?
Returning to the previous file you made using seqkit and the fx2tab subcommand, you will now be able to load this file in R. Navigate to your R console and use the file inspector in the lower right corner of your window to check for the mydata.txt file that you made above (hopefully you have not named it this). If you do not see it, make sure that you are in the correct directory.
If you do see it, you should be able to load the file into R as a dataframe. Perhaps the easiest way to do this is by using the read.table() function. Use the help function in R to see what this function does and what the syntax is. Next, load the file into a variable in R. To do this, you will need to use the read.table function but assign the output of this command to a variable. For example:
# make sure you are in R
# It is unlikely that this file name is correct
# and please don't assign it to a variable named "mydata"
# You should be able to tab-complete the name if
# you are in the correct directory, or tab complete both
# the directory and the filename
mydata <- read.table(file="data.i.made.using.seqkit.txt")
Check that you have correctly loaded the data using head(mydata) or summary(mydata).
Finally, you can plot the data. If you have used seqkit fx2tab as suggested above, your data will have four columns; the third and fourth of these are the length of the read and the quality of the read. Go ahead and plot the data in a manner you might think is informative (e.g. hist(), plot(), boxplot(), barplot(), etc.). Please think about what and how you would like to plot the data and what type of plot would be needed. Not all of the previous plotting commands are useful for this data. You can browse plotting options and methods here.
The Illumina data will have four columns. This is because of the naming scheme. Make sure you use only the relevant columnbs for plotting. The nanopore data has the expected three columns.
Portfolio Analysis
See here for brief TLDR help (or below)
-
The GC (guanine and cytosine) content of a genome (and read) is known to affect how easy it is to sequence. For example, regions of genomes that are GC-rich are often under-represented in sequencing data. One question that arises from this is whether GC content is correlated with quality scores for either Illumina or Oxford Nanopore sequencing reads. Please address this question.
You will have to generate new data on GC content and quality using the
seqkit fx2tabsubcommand (make sure you use the help command, so:seqkit fx2tab --help), and combine it withRplotting methods to answer this question. Please do this, and state whether you think there is, or is not, a link between GC content and read quality in this data, and provide your graphical analysis as evidence (you are free to present additional evidence).You may need to access specific columns of your matrix to do this analysis. If you want to do this, there are different methods. Return to the R Bootcamp lab to remind yourself.
If you decide to tell
seqkitto add a header line to your file (see option-H) then you will have to go into the file itself and edit the top line, which will probably contain a#. Remove this hash, re-save the file, and thenRshould read it properly.This analysis is part of the Portfolio Assessment:
In the practicals for weeks 4 to 12, you will encounter one question in each practical session that relates to different methods of visualisation. These will be highlighted as a “Portfolio Analysis”. For these, you will need to perform the necessary analyses and accompanying visualisations to communicate the results in a clear and aesthetically pleasing manner. You will also need to write a brief caption for each figure (100 words or less) explaining the visualisation and why you selected that particular one. Finally, you will also submit the code you have used to generate the visualisations. This code needs to be commented (for example, an average of one comment per line of code). Submit these to via Stream as a single .pdf. These should be in the order: code, followed by visualisation, followed by caption. If you have used both
terminalandRcode, please submit them together, with comments delimiting each section.
Reminder of the steps you have completed today
Getting there.
Portfolio TLDR help
TLDR summary of how to approach to Portfolio Assessment from Week 4.
- Find a way to get the GC content and any other useful info from the fastq.gz sequence files using
fx2tab - Write the output to a file; this will automatically be in tabbed column format.
- Import the data (i.e. file) into R;
read.table()is one possibility.read.tablehas a default expectation of tabbed column format - Make sure you know what the columns mean. You can rename them in the text file or using
col.names=c("sequence-name", "length", "etc")within theread.table()line. You can also rename them after having read in the data usingcolnames(mydata) <- c("sequence-name", "length", "etc"). Note that in naming variables and column / row names, spaces are highly discouraged. - Look for correlations or patterns in the data: in Chrome, How can I look for correlations in data
- Produce visualisations of your findings and explain/interpret them in a caption below the visualisaltion.
Appendix
You wanted a complete list of every sequencing platform, the read length, and the cost per Gbp?
This is from Albert Vielella
| Platform | Read length max: (paired-end*, Half of data in reads**) | Price per Gbp min: ($) |
|---|---|---|
| ILMN iSeq 100 1fcell | PE150* | 485.00 |
| ILMN MiniSeq 1fcell | PE150* | 233.33 |
| ILMN MiSeq 1fcell | PE300* | 117.97 |
| ILMN NextSeq 550 1fcell | PE150* | 43.80 |
| ILMN HiSeq 4000 2fcells | PE150* | 25.00 |
| ILMN NextSeq 1000 P1/P2 1fcell | PE150* | 30.61 |
| ILMN NextSeq 2000 P3 1fcell | PE150* | 17.30 |
| ILMN NextSeq 2000 P4 1fcell (2024) | PE150* | NA |
| ILMN NovaSeq SP 2fcells | PE250* | 10.90 |
| ILMN NovaSeq S1 2fcells | PE150* | 10.90 |
| ILMN NovaSeq S2 2fcells | PE150* | 7.97 |
| ILMN NovaSeq S4 v1.5 2fcells | PE150* | 4.84 |
| ILMN NovaSeq X & X Plus 1.5B 10B 2fcells (Feb 2023) | PE150* | 3.20 |
| ILMN NovaSeq X & X Plus 25B 2fcells (H2 2023) | PE150* | 2.00 |
| ILMN NovaSeq X & X Plus 25B 2fcells Long Reads (H2 2023) | 5000* | 13.50 |
| ElemBio AVITI 2fcell | PE150* | 5.00 |
| ElemBio AVITI 2fcell x3 pricing model | PE150* | 2.00 |
| ElemBio AVITI 2fcell 2x300 Cloudbreak (early 2023) | PE300* | 5.00 |
| ElemBio AVITI 2022 2fcell 2x75bp (Q4 2022) | PE75* | NA |
| Singular Genomics G4 F2 4fcell Standard | PE150* | 16.00 |
| Singular Genomics G4 F2 4fcell Max Read | 50 | 16.00 |
| Singular Genomics G4 F3 4fcell (Q1 2023) | PE150* | 7.4 |
| Singular Genomics Systems PX (2024) | PE150* | NA |
| PACB 8M Sequel II/IIe v2.0 HiFi 1SMRTcell | 15Kbp-18Kbp** | 43.30 |
| PACB 25M Revio HiFi 4SMRTcells (2023H1) | 15Kbp-18Kbp** | 9.95 |
| PACB Onso Short-reads (2023Q2) | PE150* | 15.00 |
| ONT Flongle 1fcell 126 channels | 20bp-2Mbp | 37.50 |
| ONT MinION Mk1b 1fcell 512 channels | 20bp-2Mbp | 13.13 |
| ONT MinION Mk1c 1fcell 512 channels | 20bp-2Mbp | 13.13 |
| ONT GridION X5 5fcells | 20bp-2Mbp | 13.13 |
| ONT P2 Solo 1fcell high duplex Kit14 | 20bp-2Mbp | 13.60 |
| ONT P2 1fcell high duplex Kit14 | 20bp-2Mbp | 13.60 |
| ONT PromethION 24fcells 10,700 channels high duplex Kit14 | 20bp-2Mbp | 10.92 |
| ONT PromethION 48fcells 10,700 channels high duplex Kit14 | 20bp-2Mbp | 10.92 |
| TMO Ion S5 510 1chip | 200-400 | 950.00 |
| TMO Ion S5 520 1chip | 200-600 | 500.00 |
| TMO Ion S5 530 1chip | 200-600 | 150.00 |
| TMO Ion Genexus GX5 1chip | 200-400 | 104.20 |
| TMO Ion S5 540 1chip | 200 | 93.30 |
| TMO Ion S5 550 1chip | 200 | 66.80 |
| MGI DNBSEQ-T10×4RS 8fcell PE100 500nm pitch | PE100* | 2.00 |
| MGI DNBSEQ-T20x2 6fcell PE100 600nm pitch | PE100* | 1.50 |
| MGI DNBSEQ-T20x2 6fcell PE150 500nm(p) (Sep 2023) | PE150* | 0.99 |
| MGI DNBSEQ-T7 4fcells | PE150* | 1.50 |
| MGI DNBSEQ-T7 4fcells x3 pricing model | PE150* | 1.30 |
| MGI DNBSEQ-G400C CoolMPS 2fcells | PE200* | 5.00 |
| MGI DNBSEQ-G400RS HotMPS 2fcells | PE100* | 5.80 |
| DNBSEQ-G99 2fcells | PE150* | NA |
| DNBSEQ-G50 FCL 1fcell | PE150* | NA |
| DNBSEQ-G50 FCS 1fcell | 100 | 720.00 |
| DNBSEQ-E25 1fcell | 25 | NA |
| DNBSEQ-E5 1fcell | 100 | NA |
| Genapsys 1M Chip | 150 | 500.00 |
| Genapsys 16M Chip | 300 | 299.00 |
| Genapsys 50M read (144M 2023?) | PE150* | NA |
| Genapsys 100M read (144M 2024?) | PE150* | NA |
| Genapsys 500M+ Chip (1B+ 2025?) | PE250* | NA |
| GeneMind FASTASeq 300 (2023?) | PE150* | NA |
| GeneMind GenoCare 1600 (2022) | 36-75 | NA |
| GeneMind Bio GenoLab M (2021) | PE150* | NA |
| QitanTech QNome-3841 | 200-2,000,000 | NA |
| QitanTech QNome-3841hex | 200-2,000,000 | NA |
| Ultima Genomics UG 100 (2flow runs) | 300 | 1.00 |
Footnotes
-
The
$PATHvariable contains places (directories) in which your computer looks for programs. These directories are listed one after the other. The computer will search these in the order they are listed until the program you requested is found (or not, then it will complain). For example, you might have a$PATHvariable that says: first look in my home directory (~/), and then in the/usr/bin/directory, and then in my friend’s directory (/friends_dir/sneaky_software_i_saved_there/). However, those are the only places the computer will look. If you want the computer to look in more places, you have to add those locations to the$PATHvariable. The$indicates that it is a variable.After installation,
condawould usually adjust your$PATHvariable by adding the directory~/miniconda3/binto your$PATHvariable – so that the programcondacan be found anytime you open a new shell, and any program thatcondainstalls will be used first because the computer will look in this directory first. However,RStudioClouddoes not recognise the addition ofcondato the path, so we add it manually. ↩ -
Paired end Illumina
.fastqfiles are often named with an internal “R1” and “R2”. This refers the “Read 1” and “Read 2” and indicates, for convencience, that the files are paired. ↩